Address
9841 Washingtonian Blvd, Suite 200 Gaithersburg, MD 20878, USA
Situation
Regulatory agencies are responsible for protecting public health across food safety, drug safety, environmental exposures, and emerging threats. The volume and complexity of data—genomic, environmental, toxicological, exposure-based—have grown exponentially.
- To manage this scientific and operational burden, the FDA has increasingly turned to predictive analytics:
- Genomic clustering for outbreak detection
- Survival and growth kinetic modeling for pathogens
- Quantitative exposure models for drugs and chemicals
- PBPK modeling for overdose prediction
- Environmental modeling for pathogen spread
Cognizance’s scientific teams have supported FDA centers by developing actionable models that reduce uncertainty and enhance regulatory decisions.
Challenge
Regulatory scientists face major barriers:
1. Highly heterogeneous datasets – Genomic, environmental, toxicological, and clinical data are difficult to harmonize.
2. Lack of standardized predictive workflows – Different FDA centers use different modeling practices, slowing adoption.
3. Limited translational models – Traditional animal and in-vitro tests often lack predictive power for real-world behavior.
4. Data overload for outbreak and exposure investigations – Investigators can be overwhelmed by thousands of genome sequences, environmental samples, or exposure scenarios.
There was a need for validated predictive analytic tools that are standardized, interpretable, and directly useful for FDA regulatory staff.
Solution
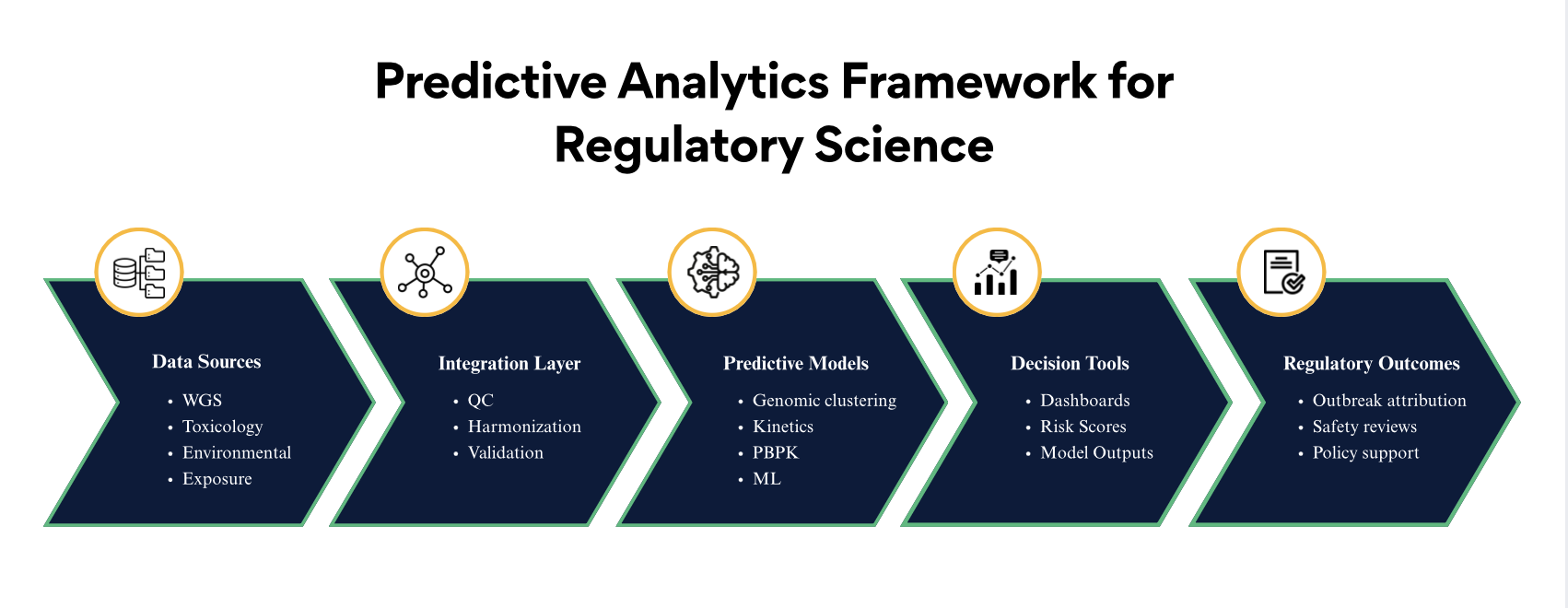
Cognizance’s scientistic teams developed a suite of predictive tools and analytics frameworks, supported by peer-reviewed publications:
1. Genomic Analytics for Food Safety
- Whole genome sequencing (WGS) pipelines for outbreak attribution
- Comparative genomics to identify strain persistence, virulence, and transmission pathways
- Automated clustering models for foodborne pathogens
Impact areas: outbreak response, contamination traceback, environmental monitoring.
2. Predictive Microbial Risk Models
- Growth/survival kinetic models on produce, mushrooms, nuts, frozen vegetables
- Transfer models for E. coli from feedlots to nearby environments
- Ecological survival models for Campylobacter in dairy systems
Impact areas: FSMA risk assessments, inspection prioritization, prevention strategies.
3. Quantitative Exposure and Toxicity Models
- PBPK models for fentanyl exposure and opioid overdose
- Toxicokinetic models for environmental stressors (ozone, PFAS, noise, particulates)
- Predictive mutagenicity modeling for nitrosamine impurities
Impact areas: drug safety evaluations, chemical risk assessments, emergency pharmacology.
4. Machine Learning & Best Practices for Predictive Systems
- ML frameworks for interpreting microphysiological systems (MPS) outputs
- Modeling guidance to standardize predictive tools across regulators
Impact areas: regulatory harmonization, model validation, cross-center consistency.
Results
The predictive analytics programs enabled:
✔ Faster outbreak attribution – WGS-based models reduced time to identify contamination sources.
✔ More accurate food safety risk assessments – Survival/growth models powered quantitative risk assessments for high-risk commodities.
✔ Improved drug safety insights – PBPK models provided mechanistic understanding of opioid exposures and countermeasure effectiveness.
✔ Enhanced environmental monitoring – Predictive models identified airborne microbial transmission pathways near agriculture facilities.
✔ Greater regulatory confidence – Standardized modeling frameworks increased reproducibility and transparency—key for regulatory adoption.
These analytics tools are now key components of ongoing FDA scientific support.
